tl;dr: It has huge advantages, but the risks are substantial enough to warrant urgent ethical, legal and policy discourse on its development and implementation.
“Most developments that start with someone saying ‘Wouldn’t it be great if …’ don’t end well”
— Maurice Chiodo
Genetic Code Swapping is a developing technology that can be used to provide complete immunity from viruses to cells by rewriting the genetic language of an organism. The technology brings with itself the potential to radically change the biosphere and enhance biosecurity with far-reaching implications like eradicating viral disease in humans altogether.
However, like most transformative technologies, Genetic Code Swapping comes with risks. In this article, I make the case that some of these risks, such as the possibility to target a custom subset of the human population with a viral pathogen, pose exceptional concern and warrant deliberation on how the technology should be developed.
I first cover some background biology. I then discuss an example of how Genetic Code Swapping has been implemented and the implications of this for future progress. Next, I describe positive use cases for and risks and problems associated with the technology. Finally, I discuss limitations of my work and give recommendations, emphasising the need for more robust ethical, policy and legal frameworks to guide the technology’s development.
Background Biology
Prokaryotic and Eukaryotic Organisms
Organisms are of two kinds: prokaryotic and eukaryotic. The former do not have a well-defined nucleus, i.e., a membranous capsule that contains the genetic material of a cell, while the latter do. Bacteria are prokaryotic while protozoa, fungi, plants and animals are eukaryotic.
DNA
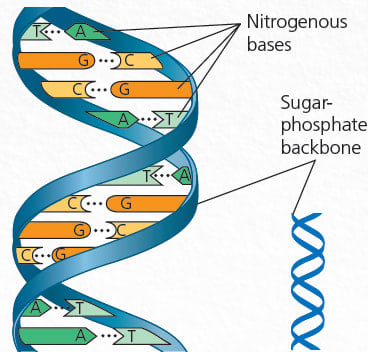
The primary genetic material in a cell is DNA, which is made up of two complementary strands of 4 types of nucleotides: A, C, T, G. The two strands are complementary in that A pairs with T and G with C, which makes DNA look like the following figure.
Transcription and Translation: From Genes to Proteins
Sections of DNA, called genes, are transcribed in a regulated manner to give mRNA, messenger RNA, a single stranded form of genetic material which is also a sequence of nucleotides. Nucleotides A, C, T, G of the DNA pair with nucleotides U, G, A, C of the mRNA respectively. In prokaryotes, mRNA is not processed, while in eukaryotes, mRNA is processed inside the nucleus then transported outside into the cytoplasm. Small units called ribosomes translate mRNA into a sequence of amino acids. A sequence of three nucleotides of mRNA constitutes what is called a codon, which either signals for a certain amino acid or signals for a stop to the translation process. A ribosome "reads” these codons to form a chain of amino acids called a polypeptide. When a polypeptide folds appropriately, it forms a protein.
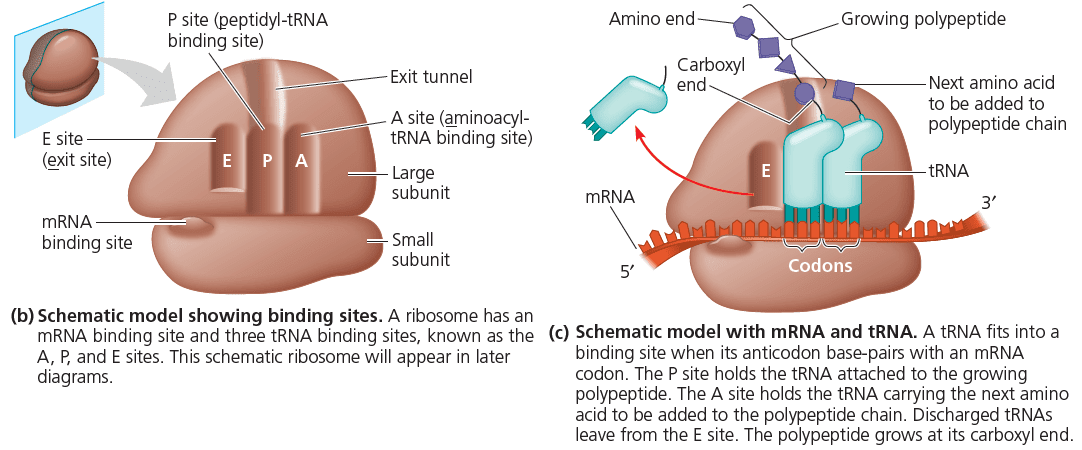
Ribosomes and tRNA: Translation Machinery
Ribosomes are made up of ribosomal proteins and rRNA, ribosomal RNA. Translation of mRNA to polypeptides happens in the following way: there are so-called tRNA, transfer RNA, that have an anticodon loop which identifies a codon on the mRNA, and they bind to a particular amino acid at another site. These tRNA (carrying amino acids with them), assemble in the ribosome and allow the entering amino acid to make a chemical bond with the chain of previous amino acids. Once this bond is formed, the tRNA bonded to the immediately previous amino acid breaks its bond and exits the ribosome. Once the last amino acid has been added to the chain, a stop codon terminates translation and the polypeptide is released. The following figure provides a helpful visualisation:
Viruses: Cellular Hijackers
Viruses consist of some genetic material (DNA or RNA) enclosed in a protein capsule. They infect cells by injecting their genetic material into the cell. The cell’s machinery may then produce viral proteins using that genetic material and create copies of the virus’s genetic material, allowing another virus to be formed.
The Genetic Code and Genetic Code Swapping
Genetic Code Swapping refers to a change in the amino acid table which maps DNA triplet codons to amino acids. The vast majority of organisms conform to the standard genetic code:
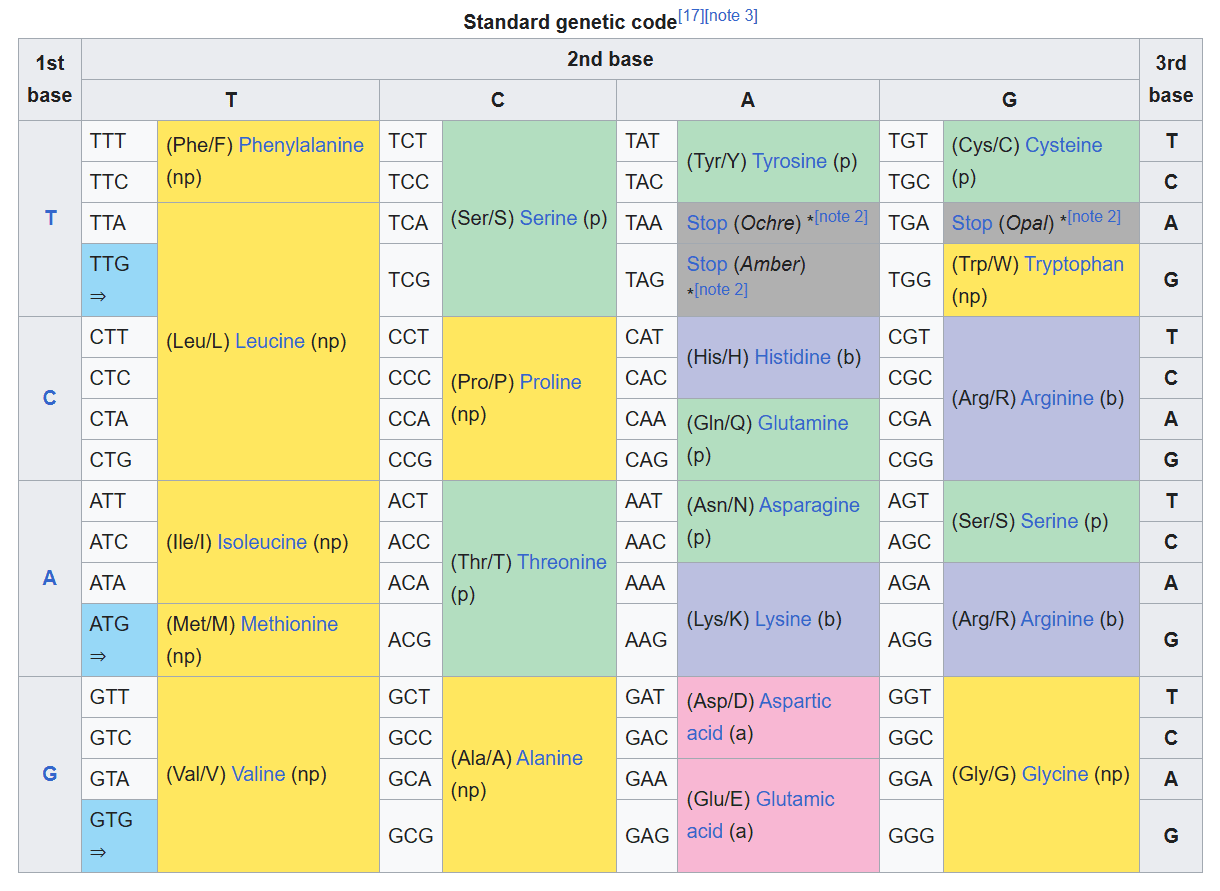
If a cell is modified such that it comes to have a swapped genetic code, then for it to continue to function normally, the cell’s DNA will also have to be modified. Given these two modifications, one would expect that viruses might not be able to infect the cell because the virus’s genes will now code for different polypeptides, i.e., yield different proteins that will no longer serve their normal functions. An article from March 2023[1] shows how researchers at the Harvard Medical School carefully engineered a code swap that conferred such immunity to E. coli, a bacterium.
Current Progress and Future of the Technology
A successful example: Syn61Δ3 and its Compressed Genome
The above-mentioned researchers used a strain of E. coli, Syn61Δ3, which was created with a ‘compressed’ genome in which all annotated instances of two serine codons, TCG and TCA (together TCR), and the TAG stop codon were replaced with synonymous alternatives, and the corresponding serine tRNA genes (serU and serT) and a release factor (RF1 (prfA)) were deleted.[1]
One might expect that the compressed genome of Syn61Δ3 would by itself confer viral resistance; however, this turns out to only partially be the case thanks to some viruses having their own tRNA genes. These viral tRNAs replace cellular tRNAs for the missing codons, and while the time taken for viral replication increases, such viruses nevertheless infect Syn61Δ3 colonies.[1]
To combat this problem, the researchers tried to repurpose TCR codons to code for leucine, an amino acid chemically very different from serine (which TCR codons usually code for). They did so by searching for tRNAs that would map TCR codons to leucine.[1]
Initially, they tried mutated versions of the E. coli leuU tRNA whose anticodon decoded TCA and TCG codons while the rest of the anticodon loop was randomized. Of the 65,536 variants tested, 2 variants were as good as wild-type tRNAs (i.e. as good enough at repurposing TCR codons to code for leucine as a usual tRNA would be at its regular job). These 2 variants were, however, not potent enough to defeat viral tRNAs completely during competition for the translation of viral mRNAs, so viruses still infected the E. coli.[1]
When the above did not work, the researchers tried anticodon-swapped variants of 13 viral leucine-coding tRNAs, since viral tRNAs tend to be more potent than bacterial tRNAs. This yielded 3 distinct tRNAs that were each as good as wild-type tRNAs and could completely block viral replication for all tested bacteriophages.[1]
The Biocontainment Strategy
Since this code-swapped E. coli strain would have a competitive advantage in natural ecosystems due to a lack of predating bacteriophages, the researchers developed a tightly biocontained version of the strain using engineered reliance of essential proteins on a human provided non-standard amino acid not present in nature. In essence, this version would be unable to survive outside a culture medium.[1]
Future Development of Genetic Code Swapping: AI Accelerated
The above method provides a blueprint for code-swapping in prokaryotic organisms. In eukaryotic organisms, code-swapping is expected to have some difficulties due to the complex nature of eukaryotic gene expression. For instance, in humans and mammals in particular, the prevalence of small non-coding RNAs called microRNAs (abbreviated miRNAs) in regulating gene expression may pose a problem. A single miRNA usually regulates the expression of more than one gene via partial binding to mRNAs. Since different mRNAs when swapped will change differently, this may lead to a miRNA that can’t be adapted to the swapped code to bind with each swapped mRNA.
It is worth noting that even in Syn61Δ3 (without the serine-leucine swap), there are 217 unintentional TCR codons (present in, among other genes, at least 4 essential genes) due to genome design errors. This causes ribosome stalling, i.e. ribosomes just stop translating because there’s no tRNA to assign an amino acid to the codon, and the whole polypeptide isn’t synthesised. In the code-swapped Syn61Δ3 variant, this leads to serine-to-leucine mistranslation, and thereby reduced fitness in nutrient-rich medium. Further, the 3 viral tRNA variants found to repurpose TCR codons to leucine each have some affinity for TCT codons, leading to serine-to-leucine mistranslation at those sites.[1][2]
In a BioRxiv preprint[2], researchers say: “Future genome design projects should focus on developing and implementing algorithms that reduce changes in promoter activity, regulatory functions, and translation rate and avoid generating unwanted cryptic promoters and translated ORFs.[3] The development of predictive models capable of forecasting transcriptional and translational effects and integrating these predictions to refactor codon composition is expected to minimize the negative fitness impact of synonymous genome recoding.”
AlphaFold 3 is a powerful AI model capable of predicting the structure and interactions of DNA, RNA, proteins and ligands.[4][5] Such a model could be made capable to provide the forecasting and integration capabilities described above and direct the desired negative fitness impact minimisation.
Positive Use Cases: Real-World Benefits of Genetic Code Swapping
Genetic Code Swapping has numerous powerful applications in various fields: global health and biosecurity, chemicals industry, agriculture, climate change, etc.
Eradicating Viral Diseases
If humans or other organisms were completely code-swapped, viruses occurring naturally would be unable to infect them.
Civilisational resilience towards viral diseases could potentially be further strengthened if sufficiently many prospective genetic codes (back-of-the-envelope calculation yields ~2.5×1085 prospective codes[6]) turned out to be possible, in addition to completely code-swapping humans being possible. If people were all made to have lots of different genetic codes, then even an engineered virus would not be able to spread rapidly and widely.
Eliminating (Accidental) Lab Leaks of Viral Pathogens[7]
If viruses and the host cells used for research in labs were both code-swapped to the same new code, then the accidental leak of such viruses would no longer affect organisms in the natural ecosystem (like humans).
Fermentation Protection[7]
Precision fermentation is the use of microbes, such as E. coli or yeast, as scalable molecule factories to produce commercially viable products such as pharmaceuticals, industrial chemicals and biofuels. For example, insulin is commercially produced by introducing the gene encoding insulin to a microbe, and huge cultures of the microbe are harvested to obtain insulin in vast quantities.
One of the big issues faced in this industry is contamination, and that contamination can be viral. It's already clear how code swapping can protect the ferment! In addition to protection, the biocontainment procedure described above can be used to ensure the swapped microorganisms won’t replace microorganisms in nature.
Genetic Code Swapping can also be used to stop microbial contamination. Viruses that would target the usual microbial contaminants can be used, because they won’t infect our factory microorganisms (since they’ve been code swapped).
N.B. The possibility of selectively targeting organisms depending on whether they are code-swapped is what constitutes the major bioweapon threat we talk about in the following section.
Orthogonal Protein Synthesis
The construction of E. coli strains that use compressed code-swapped genomes of 57 or 61 codons means that the extra 7 or 3 codons can be repurposed to encode desired synthetic proteins without affecting the normal physiology of the cell.
Transplant Immunity[7]
While code-swapping entire humans may be ethically problematic, code-swapping some tissues may be more acceptable. In the near future, we may be able to synthesise tailored tissues for transplant surgeries. These tissues would likely be designed to share the recipient’s Major Histocompatibility Complex (MHC), which is a group of genes that encode proteins and cell-surface markers involved in immune function. Ensuring the tailored tissues share the recipient’s MHC makes sure the recipient’s immune system doesn’t reject the transplant.
Such a tailored tissue can be made (effectively) immune to viral infection by performing Genetic Code Swapping on its cells. This may build to a protective chimerism that doesn't change every cell in the body, merely the ones that were getting changed anyway.
Crop Protection[7]
Viruses pose problems in agriculture. For example, the Papaya Ringspot Virus devastated the papaya farming industry. Genetically modified papaya variants resistant to the virus were developed and were farmed for human consumption as well as to create firewalls for non-genetically modified crops. Such agricultural catastrophes can be averted by code-swapping crops so they would be immune to all natural viruses.
Carbon Dioxide Capture, Biosensors, etc.
The US Department of Energy in their grant document for an E. coli genetic code-swapping (and -compression) project[8] states:
“This award will now complete the construction of a strain of the model bacterium Escherichia coli with a fully redesigned (recoded) genome that will enable safe and efficient approaches for engineering strains and pathways with diverse applications, including synthesizing biofuels and industrial chemicals from biomass or polymer waste, capturing carbon dioxide, and developing additional tools to enable further scientific discoveries. The project will also develop E. coli strains capable of synthetizing redesigned proteins using a parallel (orthogonal) protein syntheses machinery without affecting the normal physiology of the cell. Strategies to generalize those approaches will be designed to implement genome recoding and orthogonal biosynthetic capacity in other bacteria that are relevant for DOE due to their industrial or environmental importance. This project will also deliver artificial intelligence-based methods for engineering a new generation of highly sensitive biosensors that can be used for basic or applied research as well as techniques for genomic, transcriptomic, and proteomic analysis of microbial communities directly within their environment. The breadth and transformative potential of the technologies and capabilities stemming from this project will not only result in new tools useful for the research community but will also address BER’s mission by providing new knowledge on the molecular genetics of fundamental microbiological processes.”
Risks and Problems
The positive use-cases stated above are indeed transformative. The following risks and problems, however, are deeply concerning.
Targeted Code-Swapping as a Genetic Weapon
Selectively code-swapping some organisms within a population can be misused to:
- Target specifically those organisms with viral pathogens
- Target specifically not those organisms with viral pathogens
- Prevent those organisms from breeding with other organisms of the same “species”
The first two are particularly problematic in the human case because one of the deterrents to warfare/terrorism via pathogen release is the fear of the same pathogens affecting the perpetrator. If the technology is implemented in humans, then when it is done so for the first time, unequal adoption coupled with these kinds of dynamics may have negative consequences for society at large.
Biodiversity Reduction Events
Non-biocontained code-swapped organisms (especially microorganisms), thanks to their competitive advantage, may have the potential to devastate populations of non-code-swapped organisms that compete for the same resources as them. This may go beyond their own species and cause biodiversity reduction events.
Such an event might potentially be carefully engineered for the perpetrator’s gain, but what is also worrisome is that this could also come about accidentally from the leak of a non-biocontained code-swapped microorganism.
Complicates Sequence Analysis
DNA sequence analysis depends on the standard genetic code. Sufficiently many prospective genetic codes turning out to be possible would make sequence analysis a computationally far-harder task. This could be especially exacerbated by codon reassignment to arbitrary non-standard amino acids.
In a reality where organisms could be readily code-swapped, but the technology is not pervasively used among humans (i.e. not a lot of people have been code-swapped or the rate of people getting code swaps is low), a breakdown of sequence analysis efforts may allow malicious actors to engineer pandemic-potential pathogens in a lack-of-legal-clarity environment.
Reversibility of Positive Swaps
In some of the positive use-cases described above, a malicious actor could potentially reverse the positive effects of code-swapping by swapping organisms back to their previous code.
Key Limitations of My Work
- The lists of positive use cases and risks (and these limitations) are largely a function of my own limited expertise, research and imagination.
- I do not develop second-order thinking insights about how the technology may be implemented (practically speaking) as it is developed, and instead consider outcomes only in theoretical realities. For example, someone giving me feedback pointed out that having research groups change their (already comprehensive) lab safety protocols is difficult.
Recommendations + Work I’d like to see
Discuss and Expand upon Positive Use-Cases and Risks
Criticise the lists I provide here and develop more thorough ones in consultation with synthetic biology experts. My choice to avoid manufacturing risky ideas may be why my risks section is short. Brainstorm risks in some kind of safe way.
Study Possible Practical Implementations of Genetic Code Swapping
- Study the economics of involved gene engineering technologies and provider-to-consumer pathways; study how providers of the technology may be financially instituted, inherent incentive structures.
- Discuss and deliberate on societal implications in different technological, legal and policy realities.
Ensure Responsible Research Environment
It is important that organisations and individuals at the forefront of the development of the technology factor in the implications of their work and conduct their research in close consultation with ethical, policy and legal experts.
Create Technology and Craft Policy to Address Risks and Problems
“Risks and problems” include the direct risks and problems associated with Genetic Code Swapping, some of which I list above. They also include problematic implementation risks, incentive structure problems, second-order civilisational effects, etc.
Develop Legal Frameworks to Mitigate Risks and Problems
For example, the collapse of synthesis screening technologies may result in a legal vacuum regarding DNA synthesis regulation. It is important that such challenges are made aware to legal experts so that work can be done in preparation for such eventualities. Additionally, regulation governing the safe development and use of the technology must be developed.
Acknowledgements
This article was developed as my BlueDot Biosecurity Fundamentals: Pandemics Course project. I owe a significant debt to Zoheb Anjum, who both introduced me to Genetic Code Swapping and allowed me to adapt sections from his unpublished work here. I am grateful to Sofya Lebedeva, Kirsten Angeles, Seth Goodwin, Rebecca Zanini, Aditya Raj, Farrel Alfaza and Shafiq Ahmed for their advice, feedback, discussions and support that helped shape and improve this project. I also thank Akos Nyerges, who helped answer questions I had from reading his Genetic Code Swapping paper.
- ^
Nyerges, A., Vinke, S., Flynn, R. et al. A swapped genetic code prevents viral infections and gene transfer. Nature 615, 720–727 (2023). https://doi.org/10.1038/s41586-023-05824-z
- ^
Nyerges, A., Chiappino-Pepe, A., Budnik, B. et al. Synthetic genomes unveil the effects of synonymous recoding. bioRxiv 2024.06.16.599206; doi: https://doi.org/10.1101/2024.06.16.599206
- ^
Promoters are sections of DNA that promote the transcription of certain genes. The preprint, among other things, calls for genome design projects to ensure promoters don't get inserted at places they'd rather not be so that random stuff doesn't get up-regulated.
- ^
Abramson, J., Adler, J., Dunger, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024). https://doi.org/10.1038/s41586-024-07487-w
- ^
https://blog.google/technology/ai/google-deepmind-isomorphic-alphafold-3-ai-model/
- ^
(Assuming
i. only the 20 normal amino acids are encoded;
ii. at least one stop codon exists; and
iii. some codons may code for nothing)
<64 codons => 64 codons to be put in 22 buckets (20 amino acids, stop, nothing) with each bucket non-empty; 64 codons => 64 codons to be put in 21 buckets (20 amino acids, stop) with each bucket non-empty
Former by inclusion-exclusion and approximation using first few terms gives ~2.3496×1085 prospective codes and latter using same method gives ~1.5101×1084 prospective codes.
- ^
Section adapted from unpublished work by Zoheb Anjum.
- ^
https://pamspublic.science.energy.gov/WebPAMSExternal/Interface/Common/ViewPublicAbstract.aspx?rv=f15e4ef5-2b73-430f-a601-cc97245ab9a6&rtc=24&PRoleId=10

Such an interesting article that I can't wait to mull over. I can't add anything of technical value but the absolute rabbit holes I've gotten into from "hmm, I should check about diseases in dogs to keep an eye on" to "wow, mosquito borne diseases are very high" to " oh my goodness, why do I have so many papers saved on impeding the ventral nerves in mosquitos to test blood hunting mechanism inhibitions..." have nearly all converted to genetically engineered mosquitos with Ago2 gene disactivation or susceptibility to infection symptoms. The fears of genetic engineering to evade diseases by almost the flip side of making the vector susceptible strikes me with the same ethical, biological and genetic risks, plus the huge issues with bioweaponory and double use tech.
Proud of you! Great job on this whole post!