In thinking about how you might identify future pandemics by sequencing wastewater, you might have a goal of raising an alert before some fraction of people were currently infected. What you're actually able to observe, however, are sequencing reads, several steps removed from infection rates. Can we use covid data to estimate how the fraction of people currently infected with some pathogen might translate into the fraction of wastewater sequencing reads that match the pathogen?
RNA Viromics of Southern California Wastewater and Detection of SARS-CoV-2 Single-Nucleotide Variants (Rothman et al 2021) is the closest study I know in this area, though it wasn't exactly what they were trying to answer. They took many samples of municipal wastewater, extracted the RNA, filtered it to increase the fraction of the RNA corresponding to viruses, and then sequenced it.
(They also did several other things, including enriching some samples for respiratory viruses, but here I'm only looking at the unenriched data.)
They got 795M sequencing reads, 337 of which they identified as covid. This means the fraction of all reads that were covid (the "proportional abundance") was 4e-7. There's a typo in the paper where they say this is "0.0004%", but I wrote to the author to ask about it and they confirmed it was a missing zero.
How many people were infected during this period? The first tricky thing in answering this question is that the sequencing quantity wasn't uniform over this period:
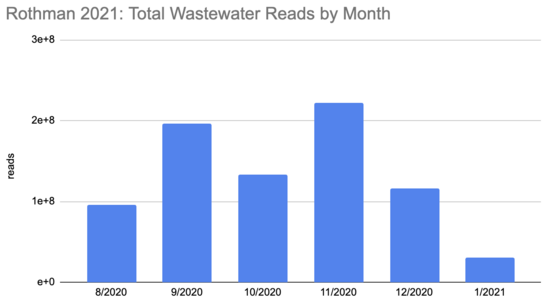
Rothman
2021 SF4_sample_metadata: xlsx
And neither were covid cases:
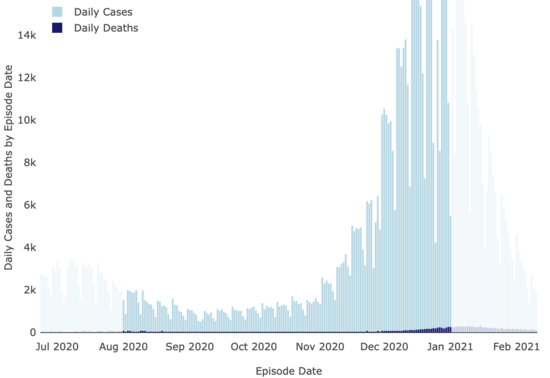
LA County Covid
Dashboard
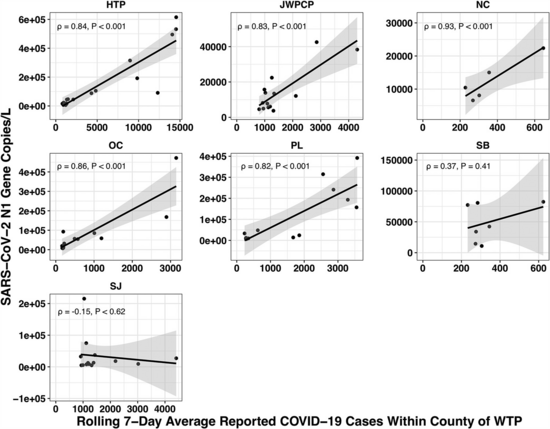
The paper accounted for the variability in confirmed cases by looking at the relationship between the number of cases in the county served by each water treatment plant and the amount of covid they measured in their samples using qPCR. Because qPCR gives you much more precise estimates for a given cost, they were able to quantify the covid composition of 85 of their samples across seven plants:
(You might be wondering: if qPCR is cheap and precise why not use it directly in looking for new pandemics? Why invest so much effort and money in sequencing? The issue with qPCR is that it's a method for checking how much of a specific sequence is present in a sample. That means you have to already know what you're looking for, and probably wouldn't be able to catch something novel.)
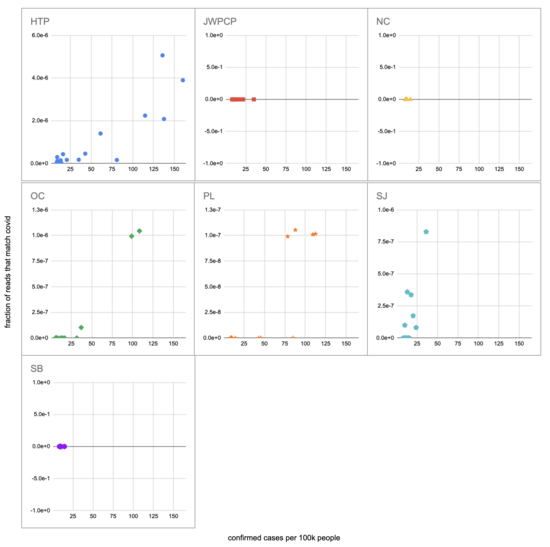
Let's recreate this chart, but quantify by the fraction of reads that match covid instead of qPCR. I identified the reads by looking for exact matches to a few of the length-40 substrings ("40-mers") of covid, and found 227 (code). I pulled confirmed cases from CSSE at Johns Hopkins University, computed a 7-day centered moving average, and joined it (code) with the sample metadata (SF4: xlsx). This gives us (sheet):
We didn't get any reads matching covid for the JWPCP, NC, or SB plants, but this isn't surprising because sequencing isn't very sensitive and they only sampled those plants early in the study when covid levels were very low.
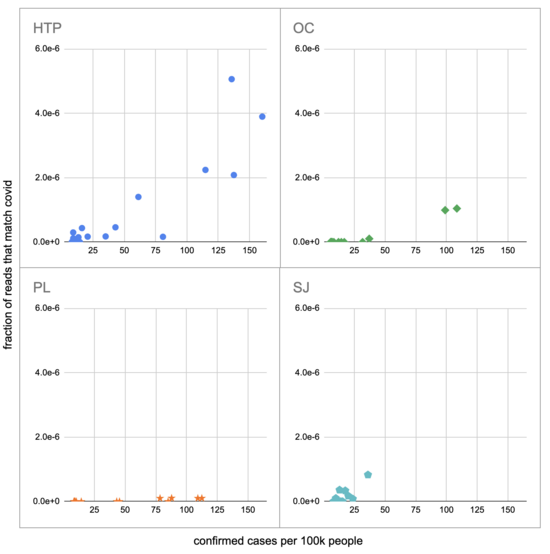
Pulling out the four plants where we do see covid, the results show a lot of variability:
The most striking contrast is between HTP, where there's a clear positive trend, and PL, where even as rates got to over 100 cases per 100k we never got more than one covid read per sample. We do know from qPCR there was covid in the water at PL, and that its concentration went up as cases went up, so it's surprising that we didn't get the number of reads we'd expect.
There's also a disagreement the other direction, where in SJ qPCR didn't show covid levels increasing with cases, but we do see an increase. This increase is not very robust, however, because it is driven entirely by a single sample, 12 covid reads on the last day (2020-11-18).
Fitting a linear model to these four, and the data as a whole, I see:
| Plant | Estimated reads at 100 cases per 100k |
|---|---|
| HTP | 2.2e-6 |
| OC | 8.6e-7 |
| PL | 7.9e-8 |
| SJ | 1.2e-6 |
| All | 1.3e-6 |
As a sanity check, let's try computing this a different way, without fancy statistics. We can compare the fraction of covid reads to the read-weighted case rate. For example, if we had:
| County | Date | Total Reads | Cases per 100k |
|---|---|---|---|
| Orange | 2020-08-12 | 2.1M | 8.4 |
| LA | 2020-08-12 | 5.8M | 20.0 |
| San Diego | 2020-08-13 | 11.0M | 8.8 |
Then the read-weighted case rate, or the mean case rate 'experienced' by a read, is:
2.1M * 8.4 + 5.8M * 20.0 + 11.0M * 8.8
---------------------------------------
2.1M + 5.8M + 11.0M
Which in this examples comes to 12.2 per 100k.
Calculating this for the full data I get (code, sheet) a read-weighted mean case rate of 33 per 100k. The paper found 4e-7 of reads were covid, so scaling by 100/33 we can add another estimate to our table:
| Plant | Estimated reads at 100 cases per 100k | ||
|---|---|---|---|
| HTP | 2.2e-6 | ||
| OC | 8.6e-7 | ||
| PL | 7.9e-8 | ||
| SJ | 1.2e-6 | ||
| All | 1.3e-6
| Read-weighting |
1.2e-6
|
|
This is quite close to what we got with linear regression, which is reassuring. Before we get too reassured, however, we should keep in mind that the PL data give much lower numbers.
Still, let's continue on with our attempt to predict a number of sequencing reads from a number of currently infected people. So far we've been using confirmed cases, and not all infections are confirmed, or even noticed. The CDC estimates that from 2020-02 to 2021-09 1:4 infections were reported. And, very roughly, the typical unvaccinated person catching covid for the first time was infected for maybe ~2w. This gives us a factor of 56 (4*14), and a new table:
| Plant | Estimated reads at 100 currently infected people per 100k | ||
|---|---|---|---|
| HTP | 3.9e-8 | ||
| OC | 1.5e-8 | ||
| PL | 1.4e-9 | ||
| SJ | 2.1e-8 | ||
| All | 2.3e-8
| Read-weighting |
2.2e-8
|
|
This is a very rough estimate: is 14 days really a typical infectious period? Is 1:4 really the fraction of infections that were confirmed during this period in the catchment area of these wastewater treatment plants? Do wastewater levels lead confirmed cases, and if so by how much? Why are the levels measured at PL 20x lower than elsewhere? Is covid similar to potential future pandemics in terms of infectious period, shedding amount, stability in wastewater, etc? Is the methodology in Rothman 2021 what you'd use if you were trying to detect new pathogens? And a single sequencing read wouldn't be enough to identify most novel pathogens, so there are additional factors I'm not getting into with later processing. Still, running through these calculations is helpful for getting a sense of how much sequencing a project like this might need.
Comment via: facebook

This is another nice piece of work looking at this problem, where cost of sequencing is factored in and they are working with pooled samples under different strategies: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4164148
The major issue with a lot of this work is how you could identify a novel pathogen and know it's a pathogen. It's not just about detecting something before it gets recognised as an outbreak, it's also about that info being actionable - if you don't know if something is a human pathogen or how it spreads and the only thing to do once an alert is raised is wait to see if clinical cases start showing up in hospitals, it's not much of an early warning system.
An alternative is to look at airplane waste, since you've got a higher probability that whatever you find came from a human, and you have a list of people to track down and see if they have mild symptoms of illness that haven't reported or asymptomatic infections. You also have info on where the disease has been imported from, which can again give you more info on what to do next.
Finally got a chance to finish reading the paper! I don't entirely understand it, though:
I think they're modeling prevalence as initially constant and then sharply transitioning to an increase of 5% per year. In thinking about infections in the cases I'm familiar with (people and wild populations) this sounds very unlike real spread, which is exponential initially (or the sum of an exponential and a constant if you're observing something new growing to exceed some background). Is their model more realistic for farmed animals, or is this just highly simplified?
It looks to me like maybe the statistical method they're using relies on this clear transition between a linear constant regime and a (very nearly) linear increasing regime, and so if the model is overly simplified (above) then their results will be too optimistic about detection. (See Figure 2 on p4 of the supplementary materials.)
They talk about being able to detect entirely novel antibiotic resistance genes (scenario 3), but I don't see anything in the paper about how they know to track a particular novel gene to see if it's an antibiotic resistance one? Is the idea that once you do realize you care about a gene you can go back and re-analyze the sequencing data you've been collecting to learn how quickly it has been spreading?
I've been working on simulating exponential growth detection: count how many times each k-mer (I've been using 40-mers) occurs on each day, and then run Poisson regression to see whether this looks like an exponential increase and how good the fit is. It works, though as discussed in this post the signal for novel viruses is likely to be extremely weak and so enough sequencing gets expensive.
Yes, that's also an important part of the problem. In some cases I think it would be clear how much of a problem it was simply from looking at it and seeing how close various parts are to matching known things (which could be automated if we're getting lots of them). But yes, in others it would be pretty hard to judge how seriously to take it.
That one seems manageable: once we recognize that something is spreading in particular areas we could use more targeted and cheaper methods, like random sampling of hospital arrivals and qPCR.
I think how it spreads is probably smaller than the other concerns. Yes, if we had more details we could make a more targeted response, but general responses like lowering thresholds for wearing PPE, ramping up PPE production, ramping up testing ability and developing cheap targeted tests, reducing some forms of non-essential activity, etc would still make sense.
As above, I think there are a bunch of things you can do aside from waiting to see if people show up in hospitals, but even then it's much cheaper to check for in hospitals if you know specifically what you're looking for.
Yes, I think airplane waste is very promising, though the statistics are likely much trickier because of the small numbers (small numbers of fliers, small number of fliers using the in-flight toilets). I'd like to see exploration of both (and also sentinel populations) to see how they compare.
Plus, depending on your sampling system, you may not have plane-level data.
(Writing for myself, not the NAO)
Thanks, great to see how sequencing wastewater may eventually help us stop a pandemic. Great work!