Summary
For pandemics that aren’t ‘stealth’ pandemics (particularly globally catastrophic pandemics):
- Reason 1: Not All 'Detections' Are Made Equal: there can be significant variation in the level of information and certainty provided by different detection modalities (e.g. wastewater surveillance vs. syndromic surveillance), and the efficacy of early detection is heavily dependent on the ability to quickly trigger an epidemiological response. Thus, the nature of the detection signal is probably an important factor affecting the time required to confirm an outbreak and take action. There should probably be a greater prioritisation of plans for public health response to different types and levels of detection signals.
- Reason 2: 'Early' Might Not Be 'Early' (or Cheap) Enough: for highly transmissible pathogens, early detection systems may only provide a lead time on the order of days to weeks compared to "naive detection" from symptomatic spread, and the costs to achieve high confidence of detection can be prohibitively expensive (on the order of billions). Improving cost-effectiveness likely requires carefully targeting surveillance to high-risk populations and locations. Methodological uncertainties make it difficult to have high levels of confidence about how valuable early detection interventions are for a range of pathogen characteristics, particularly for GCBR-level threats.
- Reason 3: Response Strategies Matter, A Lot: the cost-effectiveness of early detection is highly dependent on the feasibility and efficacy of post-detection containment measures. Factors like public compliance, strength of the detection signal, degree of pathogen spread, and contingencies around misinformation can significantly impact the success of interventions. The response strategy must be robust to uncertainty around the pathogen characteristics in the early stages of a pandemic. More work is needed to ensure readiness plans can effectively leverage early detections.
Background
I want to start this post by making two points. Firstly, I think it is worth flagging a few wins and progress in pathogen-agnostic early detection since I began thinking about this topic roughly nine months ago:
- The publication of ‘Threat Net: A Metagenomic Surveillance Network for Biothreat Detection and Early Warning’ by Sharma et al., 2024.
- The publication of ‘Towards ubiquitous metagenomic sequencing: a technology roadmap’ by Whiteford et al., 2024.
- The publication of ‘A New Paradigm for Threat Agnostic Biodetection: Biological Intelligence (BIOINT)’ by Knight and Sureka, 2024.
- The publication of the preprint, ‘Quantitatively assessing early detection strategies for mitigating COVID-19 and future pandemics’ by Liu et al., 2023.
- The Nucleic Acid Observatory continued its work, publishing several notebooks, resources, white papers, reports, and preprints and even creating a tool for simulating approaches to early detection using metagenomics.
- The UK government published its biological security strategy in June 2023, which included goals such as the establishment of a National Biosurveillance Network and the expansion of wastewater surveillance.
- The U.S. Department of Health and Human Services (HHS) announced actions the department will take following National Security Memorandum 15, signed by President Biden, including accelerating advanced detection technologies.
- The Armed Forces Health Surveillance Division’s Global Emerging Infections Surveillance branch hosted its first Next-Generation Sequencing Summit.
- Various funding opportunities for improving diagnostic technology were announced, including:
- The National Institute of Biomedical Imaging and Bioengineering’s (NIBIB) Point-of-Care Technology Research Network (POCTRN) announced funding opportunities from its centres.
- BARDA’s DRIVe project solicited abstracts for various technologies, including sequencing technologies.
- Continued advances in pathogen-identification technologies, including the very recent development of an all-in-one genetic stability assay by MilliporeSigma.
Secondly, for pandemics where a virus with an extremely long incubation period could infect most of humankind (“stealth pandemics”) before being detected, I think the prima facie case for early detection as an essential tool to mitigate GCBRs is very strong. Without intentional early detection, a symptomless pathogen could evade detection for years. Chronic HIV infection can last 10 years or longer. Given the ability to infect everyone without detection, it is plausible that manufacturing such a pathogen would be one desirable route for an omnicidal actor. Assuming we are adequately not able to prevent even one instance of a stealthy pathogen emerging, there are few viable alternatives to routine pathogen-agnostic early detection outside of a syndromic setting - such as wastewater surveillance. NAO are explicit that, “the NAO’s fundamental mission is to ensure that humanity can reliably detect stealth pandemics as soon as possible”.
I make these two points to highlight that improving early detection to mitigate the risks from GCBRs is important, tractable, and has even had a number of wins. However, I started thinking about early detection with a doe-eyed optimism about its value for mitigating GCBRs and preventing pandemics more generally. I even found that disease surveillance is amongst the most well-funded issue priorities within biosecurity. However, I think this picture masks the limitations of early detection for non-stealth pandemics; the very high degree of sensitivity of early detection interventions to government response strategies (for which there is much work to be done), and the sheer heterogeneity of the efficacy of early detection interventions across pathogens and threat models.
Better biosurveillance is a crucial part of dealing with pandemics, and I think the current level of attention early detection has received is probably warranted. However, insofar as I used to think work on early detection is both one of the safest bets for GCBR mitigation and also not very neglected, I no longer think either of these to be true. It is not clear that pathogen-agnostic early detection is amongst the very best biosecurity interventions without more work being done on standardising policies, norms, and cooperation related to countermeasures. At the same time, there are likely many holes in GCBR-related early detection efforts and much work to be done to ensure comprehensive biosurveillance in the future.
In hindsight, I don’t think any of what I have to say here is particularly groundbreaking: I think a lot of this is clear to those who’ve thought a lot about early detection already. However, I am surprised that many of these considerations weren’t more apparent in many of my earlier interactions about early detection. I was probably just in the wrong conversations. However, I also imagine I’m not the only one in this position. At the bare minimum, I hope this post encourages deeper and more widespread engagement with the challenges of ensuring early detection of pandemics.
The remainder of the post lays out some reasons for this update and some preliminary modelling conducted. However, my thinking about this is ongoing, and I’m not considerably confident about any of the takes here. I’m hoping to disprove or substantiate many of these intuitions (and more!) with more robust epidemiological modelling in the near future.
Reason #1: Not All ‘Detections’ Are Made Equal
For pathogen-agnostic early detection, we are primarily concerned with the use of technologies such as metagenomic sequencing, in which the primary result is usually a sequence of base pairs corresponding to all or part of a single DNA fragment, i.e. a read. A read might give us a string of base pairs that look like the following (in the FastQ format):
| @SEQ_ID GATTTGGGGTTCAAAGCAGTATCGATCAAATAGTAAATCCATTTGTTCAACTCACAGTTT + !''*((((***+))%%%++)(%%%%).1***-+*''))**55CCF>>>>>>CCCCCCC65 |
Where the first line is the sequence ID, the second is the sequence itself, and the fourth line encodes the quality for each base pair. By comparing these reads to established databases, bioinformaticians can attempt to identify a pathogen; identify sequences for known virulence factors, and utilise the epidemiological context to figure out if an outbreak has taken place, for example, by observing the distribution of symptomatic presentations at medical facilities. However, what we really care about is the level of information about a potential outbreak sufficient to motivate political action, and this introduces a great deal of variance in the efficacy of detection systems for catching an outbreak early enough.
The identification of a novel coronavirus was announced on 7 January 2020, but it was not until 23 January 2020 that Wuhan was shut down by Chinese authorities, and it was not until 30 January 2020 that COVID-19 was declared a public health emergency of international concern[1],[2]. Much of this delay reflects the usual epidemiological complexities of predicting whether an outbreak will lead to an epidemic or pandemic[3]; the usual complexities of motivating political institutions to act quickly[4], and time spent on further testing to ensure an appropriate level of confidence. We should also be cautious of some of the particularities of the COVID-19 reporting in China at the time[5]. However, a further implication is that this delay between a formal detection and a suitable confirmation is probably somewhat sensitive to the detection procedure itself.
Wastewater surveillance data has the disadvantage of not necessarily being connected to a patient or the identifiable spread of a pathogen. Additionally, translating viral titres in a sample to a number of cases is challenging[6]. This is less of a problem for stealth pandemics where there would be no symptoms or identifiable spread in the first place. However, we should expect that even if dynamic detection methods flag the exponential growth of an unusual pathogen, the time taken to confirm this reflects an international health emergency might be too late. Conversely, a cluster of patients showing up to a hospital with an unusual presentation of symptoms could lead to confirmation much faster. The detection modality is not the only factor that affects the level of certainty at detection. Geographic spread; the homology of the pathogen; how unusual symptoms are; the rate at which a public health body can operate, and political will are all key factors.
I suspect the nature of what constitutes a ‘detection’ is a neglected aspect in comparing difference surveillance systems, particularly taking into account local context. But more importantly, policy-oriented solutions should ideally include formal plans for how public health bodies should respond to various levels and types of sequencing data. For various surveillance modalities, lowering time-to-confirmation is an important part of lowering time-to-detection. The upshot is that the cost-effectiveness of many early detection strategies is, in part, a function of the tractability of motivating an adequate epidemiological response to detection. This will be a recurring theme.
Reason #2: ‘Early’ Might Not Be ‘Early’ (or Cheap) Enough
I’ve added some context to the ambiguities of a ‘detection’, but putting numbers behind this is important for scope-sensitive analysis. It is useful to contextualise precisely what constitutes an ‘early’ detection. In general, an outbreak that gives rise to the spread of unusual symptoms, or the spread of usual symptoms in an unusual manner, will probably be detected assuming adequate monitoring. COVID-19 was likely circulating in Hubei as early as mid-October to mid-November 2019, but it was not until a cluster of hospitalisations beginning on December 8 that initiated the testing that would eventually give rise to the confirmation of COVID-19 a month later. The ‘time-to-detection’ here is something like 50-80 days. Whilst for stealth pandemics the purpose of early detection interventions is to catch a pathogen that may not be caught until much later, if at all, most biosurveillance work exists to decrease the time-to-detection relative to this ‘naive detection that is very likely to occur anyway. This is a trivial point, but some epidemiological modelling shows why beating a naive detection in a cost-effective way may be a much harder task than we think.
By 3 January 2020, China had reported 44 cases to WHO. For simplicity’s sake, let’s take 44 hospitalisations as the number of hospitalisations required to motivate a naive detection 4 days later: even if the reality was likely a shorter level of hospitalisation required and a much longer time required to confirm this. We can also compare COVID-19 with measles:
- Measles has a reproduction rate (R0) of 15-20, versus the wild type of COVID-19’s 3.28
- Measles has an incubation period of 10 days, versus the wild type of COVID-19’s 7 days.
- Measles has an infectious period of 10-12 days, versus the wild type of COVID-19’s 5 days.
- Measles has a hospitalisation rate of 20%, versus the wild type of COVID-19’s 0.162% in England. However, I treat hospitalisations as healthcare-seeking for influenza-like illnesses (50%) which is more realistic.
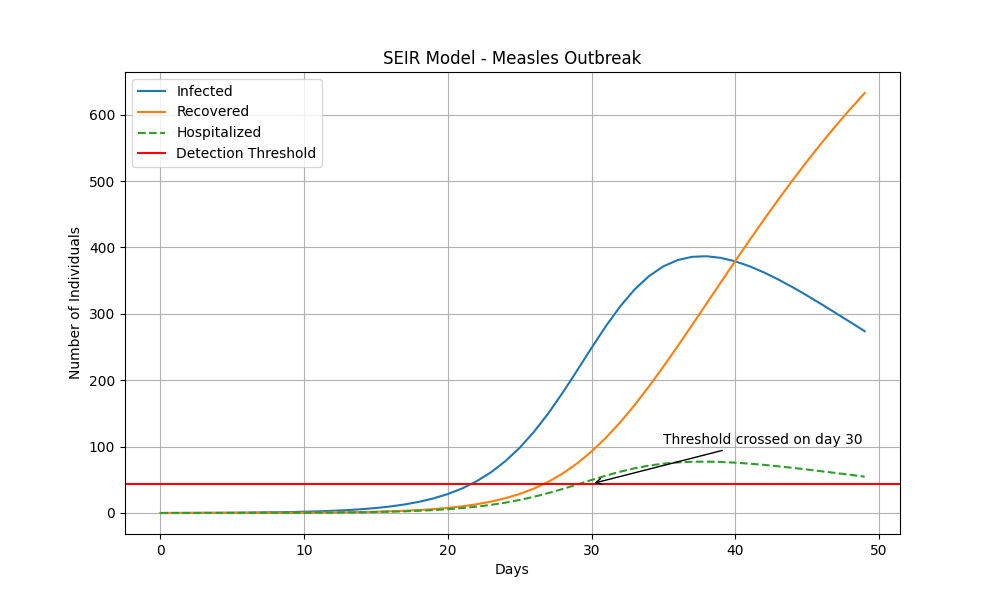
Deploying a very simple SEIR model[7], we see that measles crosses this detection threshold after 30 days:
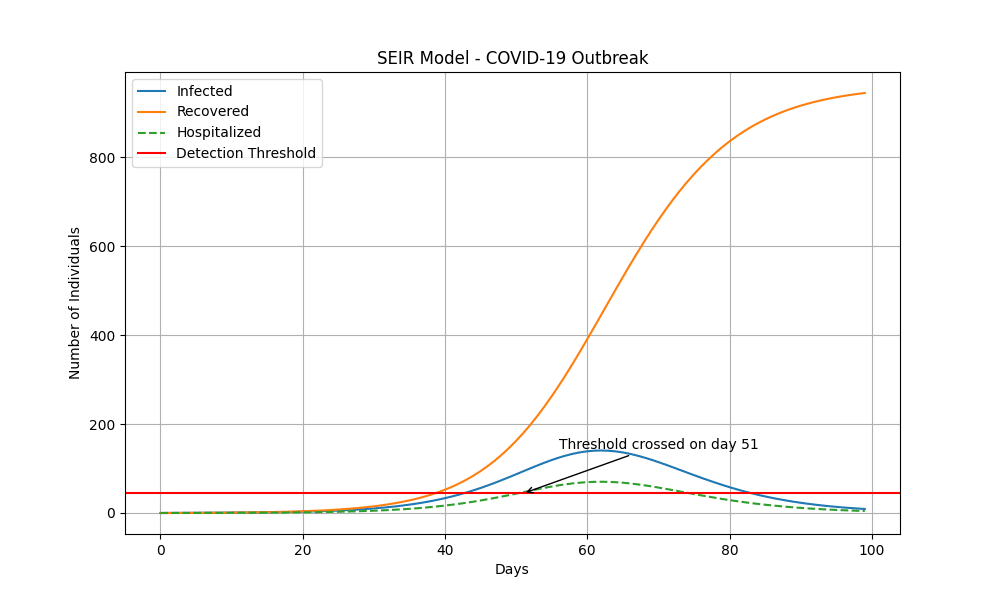
Even assuming much more generous assumptions about COVID-19 (focusing on healthcare-seeking rather than hospitalisations) than for measles, the disease does not cross this threshold until over 20 days later[8]. In practice, one would expect a measles-like pandemic to be naively detected even more quickly than this.
These figures help put into context the values we then see from publications such as Threat Net: A Metagenomic Surveillance Network for Biothreat Detection and Early Warning, which concludes devises an architecture in which 30% of the US population is covered by hospitals equipped with metagenomic sequencing capabilities, and 25% of healthcare-seekers would be referred to an emergency department where they are screened. In this proposed architecture, there would be a 95% chance of detecting a novel pathogen with the properties of SARS-COV-2 after 50 days.
To fully understand whether these figures are any good, it is useful to think about the lower bounds. The lower bound is effectively the shortest turnaround time until detection, and it is theoretically possible to get very lucky: a strange presentation of symptoms and a very high rate of hospitalisation could mean the lower bound is a little bit more than the incubation period of a pathogen, which can be less than a day for some (particularly foodborne) pathogens. Regular screening - particularly at sites like airports - could pick up a concerning pathogen within hours if lucky.
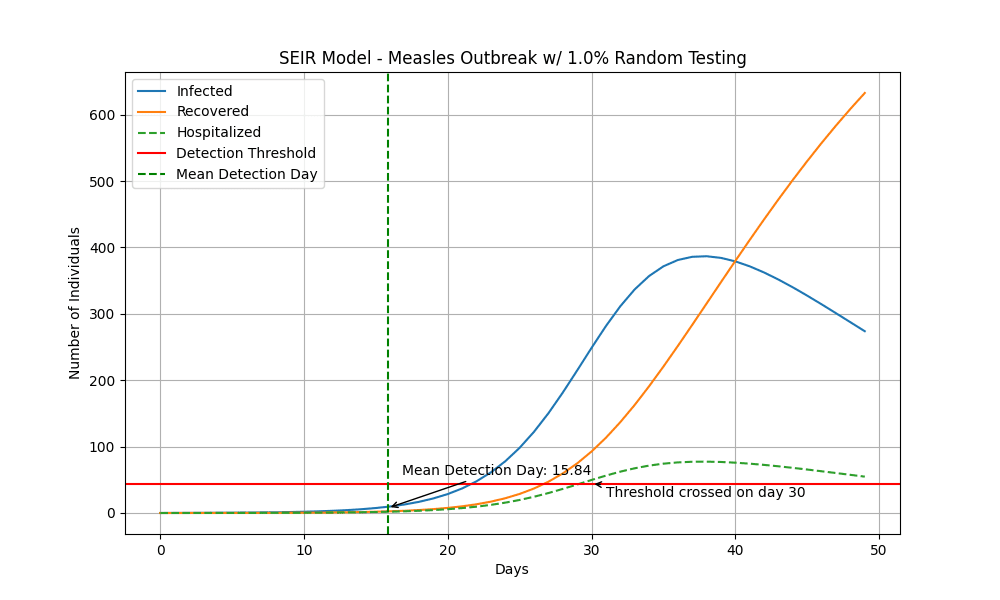
However, these cases represent getting very lucky. Another way to think about a more plausible lower bound is by considering what levels of random sampling of the population result in detecting a pathogen. With very naive modelling[9], if 1% of the population were tested every day and there was an outbreak of a novel pathogen with the properties of measles, we could plausibly half the time compared to my previous estimates.
Regularly screening 1% of the population every day is a lot. In the UK, this would be ~680,000 people per day, just over half the 1.3m people the NHS sees every day. As a very loose estimate, we can anchor on the costing provided in the proposed Threat Net architecture. Threat Net has the all-in variable cost per test at roughly $1500[10] in the US. Assuming 1 test = 1 sequencing run (e.g. so it also serves a diagnostic purpose), it would cost upwards of $372bn per year to test this many people[11]. Even assuming 10 samples can be multiplexed, £37.2bn would be close to 20% of the UK’s healthcare budget. Waiting to multiplex many more samples than this would contravene the purpose of early detection.
The specific numbers here are likely wrong, but this is meant to illustrate the broader point: in a country with functioning public health monitoring, a sufficiently transmissible pathogen might be naively detected after the index case (i.e. with no intentional early detection system) within weeks to months. A GCBR-level ‘wildfire pandemic’ could plausibly be naively detected in days. Early detection systems buy us an amount of time in the order of magnitude of days to months. However, the cost can be prohibitively expensive.
I think at least three key uncertainties and three key takeaways emerge. The first uncertainty is that it is apparent that the utility of early detection interventions is constrained heavily by a pathogen’s characteristics—especially the transmissibility, incubation period, mortality rate, and the novelty of symptoms in question. This introduces considerable heterogeneity. A pathogen with a long incubation period is poorly suited to syndromic surveillance systems, and a pathogen that has lower shedding rates in faecal material or urine is less suited to wastewater.
Even putting aside this heterogeneity, understanding the cost-effectiveness of early detection ideally requires understanding the probability distribution of pathogen characteristics that threaten us - which is not information I could not readily come across. This is possibly for good reason, but my impression is this is mostly undertheorised, particularly for GCBRs.
Secondly, scaling up early detection systems can be very expensive; and selecting better populations or sites to sample from might be a key factor in making this cost-effective. Particularly (i) populations or sites near likely sources of zoonotic transmission (e.g. reservoirs);(ii) populations or sites near potential targets for a bioterror attack, and (iii) populations that exhibit significantly higher contact rates (such as frequent flyers). A fourth category would be (iv) populations that exhibit much higher levels of viral load in the early stages of an infection (e.g. due to weaker immune systems), but I am much less confident there will be a significant early detection benefit here. However, overall, I think figuring out the right populations to sample from is an additional key uncertainty.
Of course, note that the smaller and less representative the sample, the more work post-detection might be required to reach the level of confirmation required to motivate political action. For very transmissible pathogens, the amounts of time in question we’re talking about make this a significant consideration. If we manage to pick up a measles-like novel pathogen on day 10; it would take another 7 days to gather sufficient data to initiate a countermeasure, yet on day 15 we would see enough hospitalisations to raise alarm, where the effect of early detection is then washed out.
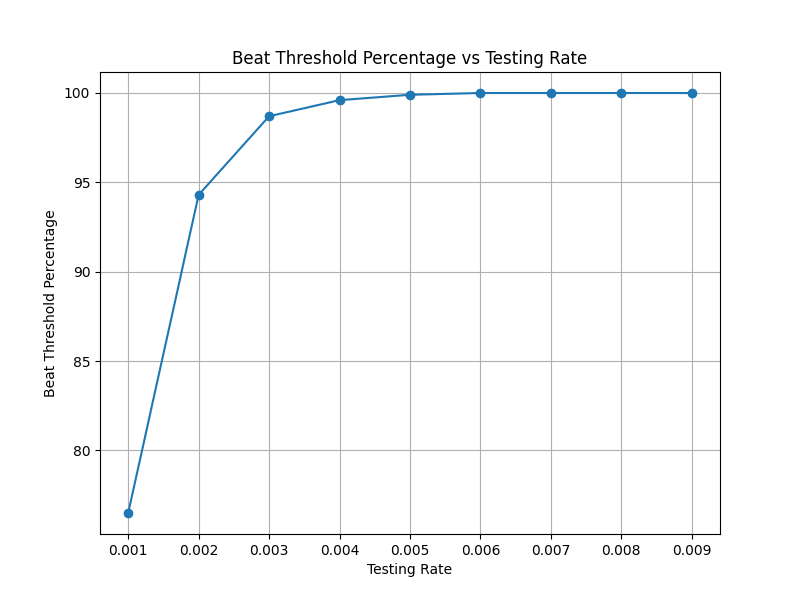
Thirdly, it is clear that the benefits of early detection are probabilistic. However, especially for those concerned about low-probability high-consequence tail risks, I think the scale in question should concern us. Ideally, we should not be content with a 5% chance of a detection system failing us for a GCBR. However, taking my measles model above, if I try and find values for what percentage of the population is tested every day such that this gives us a 99% chance of detecting the pandemic, this works out to 0.4% of the population being tested at random every day:
Once again, I note the key point here isn’t this exact number as this modelling is extremely rough. However, I do think these results are indicative of the operative points: when we are thinking through the value of early detection, we should note that the cost/feasibility burdens could plausibly be extremely large. The more transmissible the pathogen, the less time we have to catch it before it’s too late and the relative order of magnitude here could be hours to days. Not only is widespread early detection plausibly very costly, possibly in the order of magnitudes of billions to be appropriately defended against GCBRs, but we may very well not have much time to improve on it at all.
My first takeaway is a prima facie scepticism about the value of early detection for preventing GCBRs in particular compared to preventative measures - with the exception of stealthier pandemics. But of course, I have no real sense of what proportion of the threat space is constituted by stealth versus wildfire pandemics, and I am not sure there is anything close to consensus here. Secondly, much of the success of early detection in these deadlier scenarios is dependent on various factors outside the detection procedure and detection architecture itself. Sample representativeness seems to be a primary cost-saving tool and the existence of plans such that we don’t need long confirmation times is a key factor in enabling these plans. It’s been written about plenty that the ideal early detection seemingly requires a layered response to cover all our bases for pathogen types, threat models, and different geographical contexts. I would mostly draw attention to just how non-linear some of these factors could be, and we should not assume that scaling existing or planned detection systems is sufficient for protecting against GCBRs.
Finally, a number of epistemic limitations here should humble our expectations. I may be wrong here and would love to be proven wrong. But without priors over the distribution of threat models; effective modelling of the distribution of pandemic intensities[12] , and given the greater stochasticity of outbreaks in the early days of a pandemic and sheer heterogeneity across geographical contexts, it is currently difficult to assess the cost-effectiveness of early detection interventions. I don’t think this precludes us from prima facie reasoning. However, it is plausible that the unique way in which environmental surveillance seems well-suited for mitigating stealth pandemics does not generalise much further for mitigating GCBRs. Ultimately, this points towards the two-pronged conclusion of (i) fewer prima facie reasons to think early detection is a uniquely cost-effective biosecurity solution, but also (ii) resolving key questions and exploring speeding up or reducing the need for a drawn-out confirmation process are plausibly neglected aspects within early detection.
Reason #3: Response Strategies Matter, A Lot
It goes without saying that the cost-effectiveness of early detection is very sensitive to what you actually do once you have a detection and enough data to shift political will. I ultimately think this consideration is largely undertheorised, and could be much more important for the efficacy of early detection than my first impressions of the early detection space.
There’s quite a range of options one might consider:
- Pharmaceutical Interventions
- Broad-spectrum antivirals
- HEPA filters and HVAC
- Rapid vaccination deployment (e.g. 100-day vaccination program)
- Scaling up indoor far-UVC radiation
- Scaling up personal protective equipment
- Telemedicine + digital health service deployments
- Therapeutics (e.g. convalescent plasma therapy)
- Scaling up health system infrastructure
- Non-Pharmaceutical Interventions (NPIs)
- Mask and face coverings
- Social distancing
- Lockdowns
- Quarantines
- Testing, tracing, isolation
- Travel restrictions
- Public health messaging
- Better sick leave policies
- Border closures
On top of explicit interventions, it is useful to consider social behaviour changes that stem from people’s awareness there is a pandemic. This reflects reduced contacts, higher levels of hygiene, and more widespread disinfection/sanitation, etc.
COVID-19 demonstrated that divergent policy responses across nations can have divergent outcomes, and how confusing and conflicting information about interventions reduced compliance. I’ll briefly note that even though there have been a number of advancements in the literature on non-pharmaceutical interventions[13], technological advancements in pharmaceutical countermeasures[14], and improved pandemic readiness infrastructure[15], there is clearly more policy-relevant work to be done on ensuring governments have readiness plans in place. However, I think there are a few reasons to think the feasibility of implementing effective countermeasures, at the very least, is very heterogeneous:
- Compliance is probably sensitive to the detection signal: I’ve touched on the importance of ensuring the detection signal is sufficient to motivate political action, and I suspect this is additionally important for ensuring compliance in the first place. Unfortunately, there’s not a lot of precedent here, but I suspect this is true given the challenges communicating uncertainty about NPIs in the early days of COVID-19 resulting in reduced compliance. Uncertain or tentative detection signals are difficult to communicate, yet a policy of overconfidence increases the chance of being wrong.
- Intervention strength is probably sensitive to the detection signal: the logic here is ultimately the same as (1), assuming compliance loosely tracks tolerance for intervention strength. I do think this is non-obvious and there are important non-linearities. I think intervention length and interaction effects with other interventions plausibly confound this relationship.
- Interventions will probably have very heterogeneous effects based on the time-to-detection itself. This is for at least two reasons:
- Firstly, time-to-detection is a proxy for the pathogen’s transmission itself, and the more a pathogen has spread, the fewer interventions (particularly localised quarantining) are on the table to begin with. This consideration means that the intervention is further sensitive to the virulence and symptoms of the pathogen in question.
- A longer time-to-detection, insofar as this can be figured out retrospectively (which is not trivial), probably also motivates more aggressive responses in anticipation of the pathogen being more widespread.
- Contingency matters: the importance of the willingness to comply for both compliance with interventions and for which social behaviour changes we should expect means that the success of interventions is at least somewhat contingent. Much has been written, for example, about the effect of misinformation on COVID-19 compliance[16]. Political leadership, economic incentives, and social trends (such as the polarisation of response strategies) are other plausible factors. Contingency is not merely relevant for compliance, but additionally the usual contingencies that affect the expected efficacy of government interventions (such as the level of technical expertise; sufficient allocation of resources, and political incentives amongst decision-makers).
- Intervention strength, nature, and duration are sensitive to pathogen characteristics. This is firstly in obvious ways related to transmissibility and pathogenicity, e.g. a high-mortality pathogen requires more staunch social distancing. Secondly, the pre-existing level of information about the pathogen affects the ability to respond, e.g. whether it is a known pathogen; homologous to a known pathogen; genetically engineered, etc. Finally, particular aspects of its symptomatic presentation; incubation period, and viral properties can affect the nature of viable interventions.
My key takeaways are analogous to the aforementioned takeaways on earlier detection. For early detection to successfully prevent GCBRs, it need not just be the case that there are sufficient early detection systems and a suitable package of interventions. The detection signal should motivate sufficient compliance and an intervention strength; the level of spread and ability to ascertain the level of spread retrospectively must incentivise interventions that will actually work given the pathogen’s specification, and various contingencies must not get in the way to preclude compliance and effective response. I think it is unclear the extent to which these truly bottleneck interventions are useful in a GCBR. However, we should note that the relevant window of urgency in question could be a matter of days. Though an exceptionally high mortality pathogen may be sufficient to provide the exact political will required to act.
Similarly, the need for a layered early detection system is probably also mirrored by the need for layered response strategies. Additionally, just as we ideally want very high detection rates against a GCBR, the demand for interventions should be high: there is no use only defending against a single wave or single variant of an exceptionally high-mortality pathogen. Additionally, I’m additionally sceptical about the level of fidelity these considerations can be modelled ex-ante. On top of the reasons above, interventions also have complex synergistic effects; non-linear effects on transmission across a given time period for a number of reasons; very variable durations and effects on transmission between interventions yet interventions rarely happen in isolation, and, in general, the very early days of a pandemic exhibit lots of stochasticity.
Ultimately, the recurring theme recurs: we have less prima facie reason to think that the consensus about NPIs and the current rate of progress is at all adequate for enabling early detection to currently be uniquely cost-effective. Yet, work focused on compliance, strengthening detection signals, and ensuring the robustness of intervention plans to a wide range of pathogens and threat models might be additionally important pieces for ensuring early detection interventions are more cost-effective.
So What?
My takes here are tentative and not hugely confident, so I hope to continue exploring some of these ideas with more rigorous epidemiological modelling and further thinking. I think an important “so what?” here is encouraging those thinking about mitigating GCBRs to consider whether working on early detection is amongst the most important things they could be working on. My sense is there are certainly a number of combinations of pathogens, threat models, and regional contexts for which focusing on preventative strategies might be considerably more important for mitigating GCBRs. For those who are excited about working on early detection, I think there is plausibly further research here that could be useful:
- Investigating strategies to speed up post-detection confirmation processes and reduce the need for lengthy confirmations, such as pre-established response plans and thresholds.
- Exploring targeted sampling strategies that focus on high-risk populations and locations to improve the cost-effectiveness of early detection systems. My guess is there is a large space of “community-based” approaches that are neither wastewater nor syndromic surveillance and could involve technological solutions, that could be promising.
- Modelling the feasibility and efficacy of post-detection containment measures across different scenarios to identify key bottlenecks and opportunities for improvement across a wide number of conceivable scenarios.
- Researching methods to enhance the strength and credibility of early detection signals to improve public compliance and political will for rapid response.
- Further thinking about layered early detection system architectures and response strategies that are robust to a wide range of pathogen characteristics and threat models.
Ultimately, the takeaway here is not that early detection interventions are not cost-effective such that we should temper our excitement. There are a number of considerations for why one might want to push on early detection interventions independent of the above considerations. These include leveraging policy windows; positioning healthcare systems towards better biosecurity, and exploiting favourable market incentives.
Rather, the key takeaway is that the cost-effectiveness of these interventions is complex and heterogeneous. I think there is simultaneously space for highly impactful further research into some of these neglected aspects, and more careful thought about the status that early detection interventions have within EA as part of the GCBR mitigation package.
Acknowledgements
The final thoughts are my own, but most of this thinking was a group effort involving Adrian Lison, Alan Hasanic, Ieva Varanauskaite, Jo Yi Chow, Kartik Sharma, and Samuel Modée. Thinking here was part of work undertaken through Oxford Biosecurity Group. Thank you all very much for your insightful contributions.
- ^
For example, see https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7378494/
- ^
For example, see https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9114873/
- ^
For example, see https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10091135/.
- ^
For example, see https://www.tandfonline.com/doi/full/10.1080/13501763.2021.1942157
- ^
I have in mind concerns around China delaying the release of information, and China’s treatment of the purported whistleblowert Dr Li Wenliang.
- ^
I’m anchoring on this perspective from Farkas et al., 2020.
- ^
Code here (largely using Claude 3): https://github.com/ConradKunadu/Pandemic-Intervention-Modelling/blob/main/SEIR_Demo_Measles.py.
- ^
Code here (largely using Claude 3): https://github.com/ConradKunadu/Pandemic-Intervention-Modelling/blob/main/SEIR_Demo_COVID.py
- ^
Code here (largely using Claude 3): https://github.com/ConradKunadu/Pandemic-Intervention-Modelling/blob/main/SEIR_Demo_Random_Sampling.py
- ^
Data and code for Threat Net can be found here. In this paper, their total cost for deploying a surveillance system for 30% of $658.1 million to operate annually independent of installation costs. In Threat Net, the daily number of tests for a US population of 331,893,745 is 1204. Therefore the annual number of tests is 365 * 1204, or 439,460 tests. Assuming 1 test = 1 run, the all-in variable cost per test is the annual cost ($658,100,000.00) / number of tests (439,460), or roughly $1500.
- ^
$1500 * 365 * 680,000 = $372.3bn.
- ^
Marani et al., 2021 is most likely the best work here, however I suspect the generalised pareto distribution used to model extreme pandemics becomes highly inaccurate at a GCBR level of intensity.
- ^
- ^
- ^
Such as the UK’s Biological Security Strategy and the launch of organisations such as CEPI.
- ^
For example, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9114791/, https://misinforeview.hks.harvard.edu/article/feeling-disinformed-lowers-compliance-with-covid-19-guidelines-evidence-from-the-us-uk-netherlands-and-germany/, https://www.mdpi.com/1660-4601/18/22/12266, and https://www.frontiersin.org/journals/psychology/articles/10.3389/fpsyg.2020.567905/full.

I work on the Nucleic Acid Observatory at SecureBio, but in this comment I'm speaking only for myself.
Thanks for writing this, and thanks @mike_mclaren for sending it to me! I wish I'd happened across it when you'd posted it [1] and could have given more timely feedback!
At a high level I think this is very reasonable, especially sections #1 and #3: it's very important that you can turn "here is a collection of highly suspect sequencing reads" into a well-calibrated and trusted "here is how concerned we should be", and it's also very important that you can then turn that into "and here's what we will do". I've seen scattered work on both of these, but I think these are key areas for additional work.
On section #2, which is closer to what I've been thinking about, there were some places where I wanted to push back or add additional information. A few point-by-point replies:
While there is a lot of spending on surveillance, this is overwhelmingly allocated to tracking known threats: how prevalent are SARS-CoV-2, Influenza, RSV, etc. I'm not aware of anyone who has deployed a system capable of alerting on a novel pathogen. While (in my work at the NAO) I'd like to change this, I do still think it's quite neglected for now.
This is definitely a real drawback. On the other hand, once your wastewater-based initial-detection system flags a pathogen you have what you need to create a cheap PCR-based diagnostic, which you can then deploy widely to identify infected individuals (to see how they're doing, if they need treatment, and understand the effect the pathogen is having in their bodies, and also to limit spread). This could let you identify many more infected individuals than if you plan was for your initial-detection system to also double as your identification-of-individuals system. But the need for follow-up work ties into the rest of your post, which is that initial detection is only one piece, and if you don't have follow-ups ready to go you'll lose many valuable doubling periods fumbling the next steps.
Strong agree!
While I agree that daily screening of 1% of the UK's population would be very expensive, I think the main cost (unless you're doing wastewater) is in collecting the samples. That's just a massive endeavor! If you get sampling down to $1 person it's still $250M/y (68M people × 1% × 365) in sample collection alone. This range might be possible if you somehow just made it part of the culture (ex: every school child provides a sample every morning) but would be a huge lift.
But I think your estimates for the cost are otherwise too high. The main thing is that if you do pooled testing you can cost-effectively sample hundreds of thousands of people at once. This isn't "multiplexing" because you're not adding barcodes (or something else) that would allow you to map sequencing reads back to specific sampled people. For example, say you're doing pooled nasal swabs, and you're sampling a large enough number of people that you'll likely get some high viral load folks. Then the average contribution of a sick person is a relative abundance of maybe 2% (rough analysis; @slg and @Will Bradshaw should have a much more thorough treatment of this out any day now). If you run a NovaSeq X 25B (which gets you 25B sequencing read pairs) on a massively pooled sample and 1:100,000 people are currently shedding, you'll get about 5k (2% ÷ 10,000 × 25B) read pairs matching the virus, which is likely (again, hoping to get something out publicly about this soon!) enough to flag it. The list price for the flow cell for a 25B sequencing run is $16k, which is perhaps half the all-in cost. Which means if you did daily NovaSeq X 25B runs you'd be spending $10-$20M/y ($16k × 2 × 365) on the sequencing.
This is all very ballpark, but mostly is to say that if you're talking about directly sampling that many people the main cost is in the collection.
This is quite different if you're using wastewater to avoid needing to sample that many people, but then your relative abundances are far lower and you need much deeper sequencing (expensive!) to get anywhere close to detection at 1:100,000.
I was briefly quite excited about this direction, but a major issue here is that these populations are, because of their vulnerability, somewhat epidemiologically isolated: they're probably not close to the first people getting sick in anywhere.
That seems too strong, at least in theory. We should evaluate the marginal detection effort in terms of how much it reduces risk for the cost. For example, a detection system that was very cheap to deploy and only flagged a pandemic 30% of the time could be well worth deploying even though it most likely doesn't help. And perhaps scaling it up to 90% probability of detection would remain cost effective, but getting it up to 99% would be massively more expensive for diminishing returns and so doesn't work out. On the other hand, political considerations might come into play: a system that on average doesn't work could be a hard sell even if it's relatively cheap.
I also don't expect work focused on initial-detection to help much outside of stealth pandemics. I see my work at the NAO as almost entirely valuable in proportion to the likelihood that someone creates (or perhaps would create, if not deterred by the likelihood of it being flagged) a stealth pandemic.
That said, there is one other situation I've seen bounced around where I think this kind of monitoring could be valuable outside of stealth scenarios: when we know there's something happening in a region inside a country we have very poor relations with. In that case if we had a system of ongoing monitoring we could pivot it to populations enriched for contact with that region and (ideally) ramp it up.
[1] A few weeks ago I happened to go through and changed my settings to give a big boost to any biosecurity-tagged posts, so I think if this came through again I'd see it.
Excellent post; I did not read it carefully enough to evaluate much of the details, but these are all things we are concerned with at the Nucleic Acid Observatory and I think your three "Reasons" is a great breakdown of the core issues.
Thank you for writing this article! As a complete newcomer to pandemic preparedness at large, I found this extremely useful and a great example of work that surfaces and questions often unstated assumptions.
Although I don't have enough expertise to provide much meaningful feedback, I did want to bring up some thoughts I had regarding your arguments in Reason 2. Your 44 hospitalizations threshold in the numerical examples strikes me as reasonable, but it does also seem to me that the metagenomic sequencing of COVID-19 was related ― if not a critical precondition ― to China confirming its finding of a novel pathogen (source). I recognize that the early detection interventions you are calling into question here may be more of the form of mass/representative sampling programs, but it seems plausible to me that merely having the means to isolate and sequence a pathogen near the site of the outbreak in question could substantially affect time to confirmation.
My prior is that China is likely quite capable in that regard, but other countries may have fewer capabilities. All this to say that investing in more conventional metagenomic sequencing capacity in "sequencing deserts" could still be very cost effective. But note that this is all conjectural; I don't know anything about the distribution of sequencing capacity, nor even what it takes to identify, isolate and sequence a pathogen.
Thanks again for this brilliant piece!
Thank you for writing out this argument! I had a quick question about #2. The earlier a pandemic would be caught by naive screening, the quicker its spread is likely to be. So despite the fact that early detection might buy less time, it might still buy plenty of value because each doubling of transmission occurs so quickly.
This still depends on mitigating the concerns you raised in #1 and #3, though.